Among natural biopolymers, DNA and RNA possess the unique ability to self-assemble into a unique double-stranded helix form in ionic media, based on a combination of Hydrogen bonding of bases, π−π stacking of base pairs, and electrostatic stabilization of the phosphate backbone. The stable DNA based helix, with a diameter of ~2 nm and persistence length of ~50 nm, has long been used as the sole building block for engineering 2D and 3D nanostructures that are used to pattern nanoparticles and biomolecules on solid surfaces. DNA has also been linked to rigid and flexible (dendritic) organic molecules, polymers, metal complexes, and nanoparticles to be used for highly selective DNA detection and to direct the assembly of those particles and complexes in a predesigned fashion for electronic applications.
The assembly of organic-DNA hybrids into nanostructures in aqueous media has generally been explored using three basic parameter groups:
1) The number of single strand (ss) DNA chains that are attached to the organic “core” fragment,
2) the orientation, geometry, and concentration of these strands, and
3) the type (Na+, K+, Ca+2, Mg+2, etc.) and concentration of salts.
Surprisingly, the role of the organic core in the assembly of nanoscale organic-DNA structures has rarely been investigated even though the core fragment can often be quite hydrophobic and should have significant interactions with the bases/base pairs in ionic media. Indeed, the hydrophobic interaction of rigid organic groups such as stilbenes and anthraquinones with DNA bases or base pairs has been utilized to stabilize small duplex DNA-detection probes to improve their target affinity and specificity. Given these precedents, it is surprising that only a few studies to date have examined the roles that hydrophobic interactions play in the assembly of organic-DNA building blocks into larger structures. Notably, perylene diimide (PDI)-DNA hybrids have been found to form both hairpin dimers based on double helix DNA assembly as well as larger supramolecular oligomers involving hydrophobic interactions of the PDI cores with each other via hydrophobic stacking.

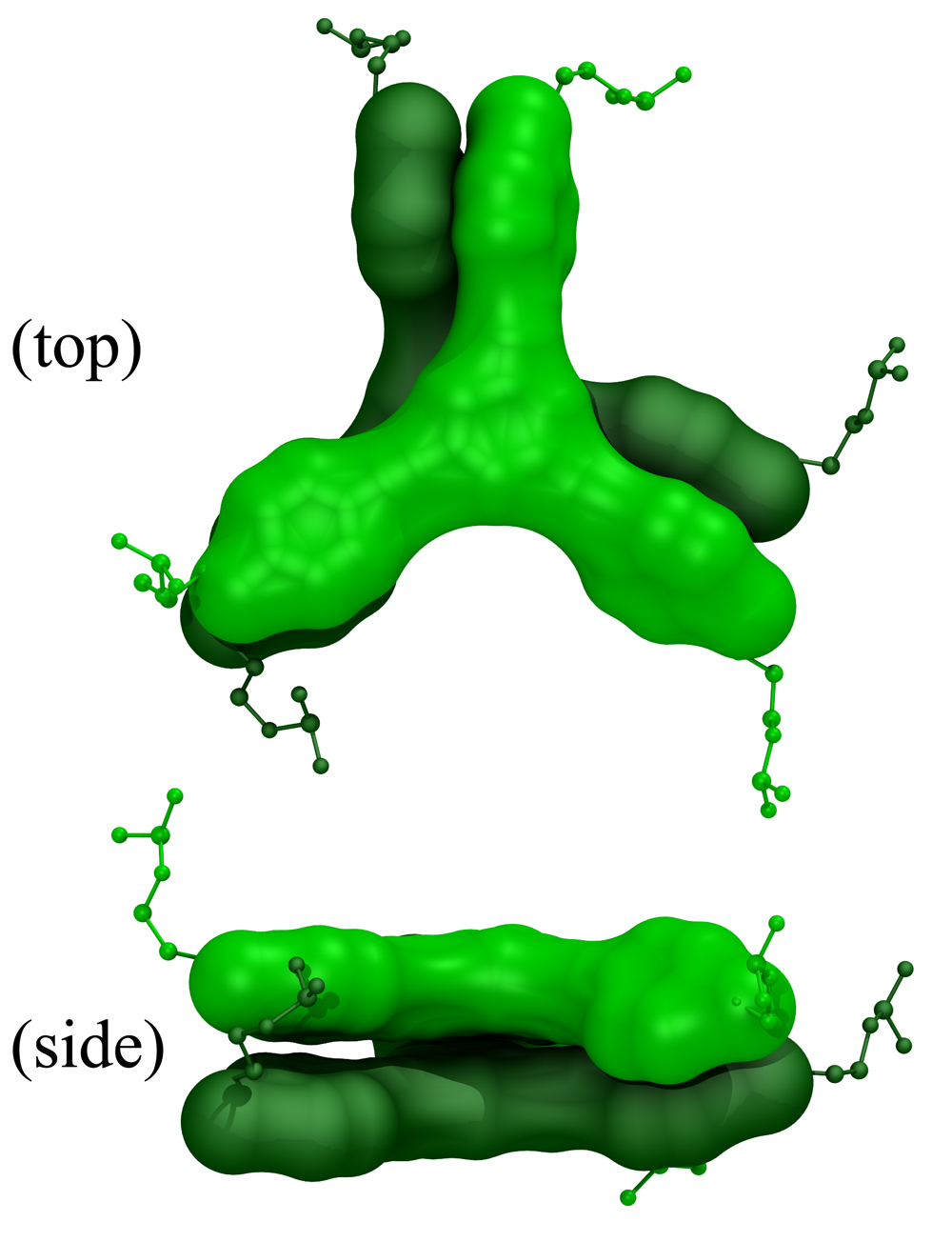
In collaboration with Nguyen Lab, we studied the effects of organic hydrophobic cores in the assembly of rigid small molecule-DNA hybrid nanomaterials into caged structures that have the potentials to be used as next generation drugs in gene and cancer therapy. It was observed that when organic cores were used in the design of DNA-hybrid materials, the assembly of DNA-hybrids was not happening in some systems. There was no clear theoretical explanation for this outcome. We provided a novel interpretation to this phenomenon. The experimental results and computational simulations showed that the final nanostructures assemble in aqueous environments such as to minimize exposure of the hydrophobic surfaces of the organic cores to solvent. These hydrophobic interactions are greatly influenced by the incorporation of multiple non-complementary deoxythymidine (T) spacers between the core and the DNA duplex, as the solvent-accessible surface area (SASA) of the hydrophobic cores is greatly reduced when these spacers are wrapped around the cores. However, when exposure of the hydrophobic surfaces of the cores could not be properly minimized in a dimer conformation, an ill defined network was forming. The results brought hydrophobic interactions into light as a new parameter that must be seriously considered in the design of DNA-hybrid materials. Furthermore, we provided novel interpretation to the directional roles of hydrophobic cores in the assembly process of DNA-hybrid nanomaterials.